Our goal is to understand how cells integrate molecular signals to build, shape and maintain different tissues across an organismal lifespan
The architecture, homeostasis and function of the organs is defined at the cellular level by the integration of a myriad of molecular signals. In our lab, we study how stem cells, progenitor cells, and somatic cells integrate microenvironmental signals, as well as how the underlying cascades define cellular identity, behaviour and resilience. We focus on unravelling molecular mechanisms in signal transduction, specially in the context of WNT signalling (See our consortium), DNA replication and damage, and chromosome segregation fidelity, which misregulation often leads to disease, notably cancer. Further, we aim at understanding how different environmental signals, including contaminants of emerging concern, impact these cellular and physiological processes.
To address these questions, we integrate cutting edge live cell imaging, single-cell genome, transcriptome and replication sequencing (scG&T-seq, scEdU-seq), and genome editing techniques with detailed molecular analyses in 2D/3D stem cell models of development, tissue homeostasis, ageing and cancer, including organoids, mouse models, and primary or immortalised cancer cell lines.
You can learn more about our research institute in Heidelberg here. We are also opening a new lab at the University of the Basque Country in January 2026. We search for motivated students and researchers. You can check how yo apply here.
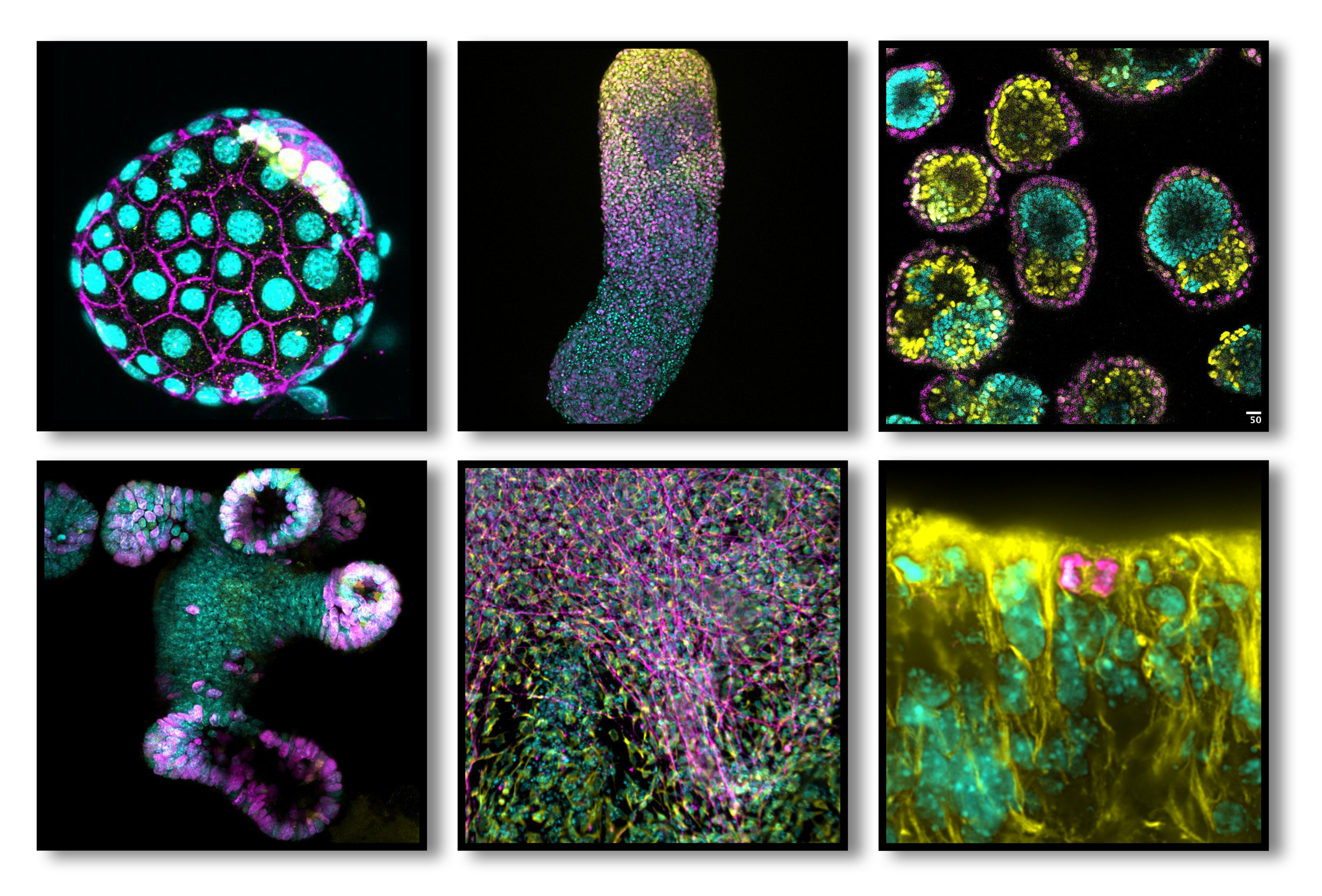
Figure 1: Biological models used in the lab. We investigate molecular mechanisms driving mammalian development and tissue homeostasis using mouse blastocysts (upper left), 3D stem cell models such as human and mouse gastruloids (upper middle), and mouse EiTiX-embryoids (upper right), mouse mammary gland and intestinal organoids (lower left), as well as in vitro generated human embryonic lineages (cortical neurons, lower middle), together with mouse models for development (developing brain; lower right), tissue homeostasis, ageing or cancer (including patient-derived organoids).

Lab members in 2023 – 2024